
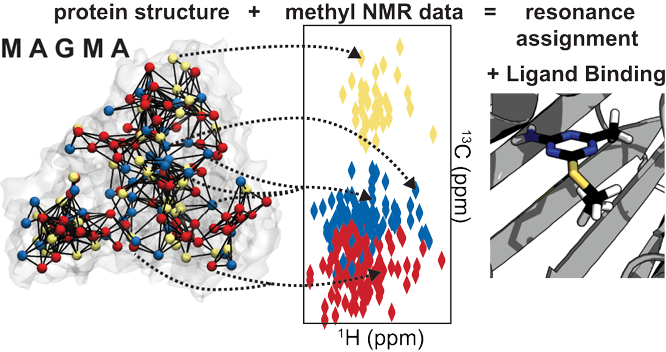
MAGMA is software for assigning methyl resonances using through space NMR data (e.g NOE for solution-state or DREAM for solid-state) using a known structure of a protein. The algorithm uses graph theory to perform an exact search of all possible assignment solutions, returning a result that should include all possible assignments each given residue, given the supplied NOEs. Accuracy on the benchmark is very high: we have a 100% success rate in confident (1 option assignments) in giving the correct assignment in our benchmark.
Details can be found in 'Pritisanac et al. JACS 2017' DOI: 10.1021/jacs.6b11358.
The version 1.2.3 for download is compiled for linux and mac. Be sure to look at 'readme.pdf' first, to walk you through running the program on the example data. Any technical inquiries, problems, or suggestions can be addressed to Prof. Andrew Baldwin at andrew.baldwin@chem.ox.ac.uk.
Software ReadMe.pdf
Please use the form below to enter your email address to receive a download link. Included are benchmark examples to use as templates, and readme files describing how to setup the program.


